Staff profile
Professor Ehmke Pohl
Professor
Co-Director and Durham Lead of the EPSRC MoSMed CDT

| Affiliation | Telephone |
|---|---|
| Professor in the Department of Chemistry | +44 (0) 191 33 43619 |
| Professor in the Department of Biosciences | +44 (0) 191 33 43619 |
| Co-Director in the Biophysical Sciences Institute | |
| Biophysical Sciences Institute Executive Board in the Biophysical Sciences Institute |
Biography
Research Interest
The overall goal of the group is to determine the 3-D structures of proteins with relevance to biomedicine or biotechnology. Our primary technique is protein crystallography complemented by a wide range of biophysical techniques ranging from small angle X-ray scattering and electron microscopy to isothermal titration calorimetry and thermal shift assays.
Virus-X: Viral Metagenomics for Innovation value

In this EU Horizon2020 funded project a consortium of private companies, led by the Icelandic biotech company Prokazyme (http://prokazyme.com/) and public researchers. will mine the unexplored genomic diversity if bacteriophages from extreme environments. Starting from a massive Next-generation sequencing effort, the goal is to discover new enzymes with novel properties for biotechnological applications. Our group is responsible for the biophysical and structural characterisation of proteins with a specific focus on DNA/RNA binding and processing enzymes.
Structure and mechanism of transcriptional regulator proteins
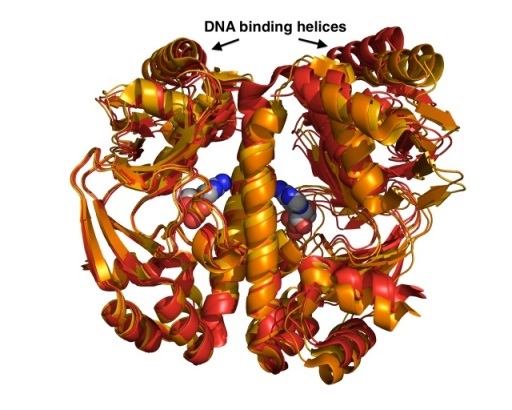
The viability of all bacteria depends on their quick adaptation to the ever-changing environment. In many cases, gene-expression in response to internal and/or external signals is controlled by transcriptional regulators. Using a combination of crystallographic, biophysical and computational tools we investigate ligand binding and the mechanism of DNA recognition. We have solved a number of crystal structures of the Catabolite Activator Protein (CAP) from E. coli and its closely related homologue GlxR from C. glutamicum to dissect structural and dynamic contributions of allosteric ligand and DNA binding. (Rodger, T.L. et al. Plos Biol (2013) e1001651; Townsend, P.D. PLoSOne (2014) e113265).
Thermal Shift Assays for protein crystallography
Thermal Shift Assays (TSA) also known as Differential Scanning Fluorimetry or Thermofluor assays are based on the change of fluorescence signal of a chemical probe. This assay can easily be performed using SYPRO orange or a related dye and a standard quantitative RT-PCR instrument. Using SBS 96-well plates the protein-dye mix is slowly heated while the fluorescence signal is monitored. When the protein chain unfolds the hydrophobic core becomes exposed and the fluorescence signal increases until complete denaturation. We have developed new screens in 96-well plate format to investigate protein stability and aid crystallization efforts. Data analysis and interpretation is automatically done by a freely available program NAMI, which reads the raw data, calculates melting temperature and displays the results in the form of color-coded table (Groftehauge, M.K., Hajizadeh, N.R., Swann, M.J., Pohl, E., (2015) Acat Cryst D71:36-44).
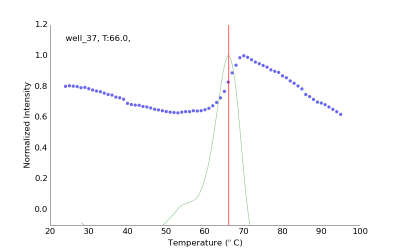
Experimental fluorescence data from one representative well (blue dots), first derivative and calculated Tm of 66 ˚C.
The program NAMI can be downloaded here, and the Durham screens will soon be available via Molecular Dimensions (http://www.moleculardimensions.com/)
Miscellaneous projects
Additional projects in the group are focused on the structure-based design of new ligands, probes and inhibitor inlcuding
- Investigating the structure and function of proteins involved in the sphingolipid biosynthesis from pathogenic protozoa in collaboration with Dr. P.W. Denny (Durham)
- Design of new chemical probes targeting the Retionic Acid Receptors (RAR) proteins with Prof. A. Whiting (Durham) and High Force Research (http://www.highforceresearch.com/)
- Structure-based design of Tuberculosis booster drugs in collaboration with the Dr. JC Cole form Cambridge Crystallographic Data Center (http://www.ccdc.cam.ac.uk/) and Dr. A.R. Baulard, Pasteur Institute, Lille.
- Development of new inhibitors for Multi-herbicide resistant weeds in collaboration with Professor P.G. Steel (Durham), Prof. R. Edwards (Newcastle) and Syngenta (https://www.syngenta.co.uk/)
Protein Crystallography Laboratory
Our group is located in the joined Biophysical Sciences Institute (BSI) laboratories in the Chemistry Department of Durham University. The protein crystallography laboratory has full access to state-of the art protein production and purification equipment (Harbiger Bioreactor, AKTA Explorer, AKTA Pure), biophysical characterization (CD, TSA, FA, ITC, SPR and MS) and crystal structure determination (Innovadyne Screenmaker, Bruker MicroStar, Bruker D8).
Acknowledgments
Financial support has been or is currently generously provided by the EPSRC, BBSRC, MRC, The Royal Society, the Wellcome Trust, the British Society for Antimicrobial Chemotherapy and Horizon2020.
Research interests
- Protein Crystallography
- DNA-Binding Proteins
Esteem Indicators
- 2017: Christopherson/Knott Foundation Fellow of the IAS Durham:
- Chair of the Programme Advisory Committee for Structural Biology at the MAXIV Synchrotron, Sweden:
Publications
Journal Article
- High-throughput investigation of macromolecular interactions for drug development using spectral shift technologyHunter, C. E., Pohl, E., & Freitag-Pohl, S. (2025). High-throughput investigation of macromolecular interactions for drug development using spectral shift technology. Biophysical Reviews. Advance online publication. https://doi.org/10.1007/s12551-025-01359-x
- Recent computational advances in the identification of cryptic binding sites for drug discoveryGašparíková, D., Chikhale, R., Cole, J., & Pohl, E. (2025). Recent computational advances in the identification of cryptic binding sites for drug discovery. Bioinformatics Advances, 5(1), Article vbaf156. https://doi.org/10.1093/bioadv/vbaf156
- Synthetic Retinoids for the Modulation of Genomic and Nongenomic Processes in Neurodegenerative DiseasesButler, A. M., Chisholm, D. R., Tomlinson, C. W., Khatib, T., Clark, J., Wan, S., Coveney, P. V., Greig, I. R., McCaffery, P., Pohl, E., & Whiting, A. (2025). Synthetic Retinoids for the Modulation of Genomic and Nongenomic Processes in Neurodegenerative Diseases. ACS Omega, 10(22), 23709–23738. https://doi.org/10.1021/acsomega.5c00934
- Profiling Serine Hydrolases in the Leishmania Host‐Pathogen Interactome Using Cell‐Permeable Activity‐Based Fluorophosphonate ProbesIsern, J. A., Porta, E. O., Kalesh, K., Koutsogiannis, Z., Cazzola, D., Pohl, E., Denny, P., & Steel, P. G. (2025). Profiling Serine Hydrolases in the Leishmania Host‐Pathogen Interactome Using Cell‐Permeable Activity‐Based Fluorophosphonate Probes. ChemBioChem, 26(10), Article e202500160. https://doi.org/10.1002/cbic.202500160
- Novel metal sites revealed by spectroscopic and structural characterization of the ferric uptake regulator from Acidithiobacillus ferrooxidansArgandoña, Y., Olivos, A., Obando, P., Imas, F., Pohl, E., Quatrini, R., & Arenas-Salinas, M. (2025). Novel metal sites revealed by spectroscopic and structural characterization of the ferric uptake regulator from Acidithiobacillus ferrooxidans. Computational and Structural Biotechnology Journal, 27, 765-777. https://doi.org/10.1016/j.csbj.2025.02.017
- Structural requirements for the specific binding of CRABP2 to cyclin D3.Pastok, M. W., Tomlinson, C. W. E., Turberville, S., Baslé, A., Butler, A. M., Noble, M. E. M., Endicott, J. A., Pohl, E., & Tatum, N. J. (2024). Structural requirements for the specific binding of CRABP2 to cyclin D3. Structure, 32(12), Article S0969-2126(24)00389-7. https://doi.org/10.1016/j.str.2024.09.020
- Toxoplasma ceramide synthases: Gene duplication, functional divergence, and roles in parasite fitness.Koutsogiannis, Z., Mina, J. G., Albus, C. A., Kol, M. A., Holthuis, J. C. M., Pohl, E., & Denny, P. W. (2023). Toxoplasma ceramide synthases: Gene duplication, functional divergence, and roles in parasite fitness. FASEB Journal, 37(11), Article e23229. https://doi.org/10.1096/fj.202201603RRR
- Independent Membrane Binding Properties of the Caspase Generated Fragments of the Beaded Filament Structural Protein 1 (BFSP1) Involves an Amphipathic HelixJarrin, M., Kalligeraki, A. A., Uwineza, A., Cawood, C. S., Brown, A. P., Ward, E. N., Le, K., Freitag-Pohl, S., Pohl, E., Kiss, B., Tapodi, A., & Quinlan, R. A. (2023). Independent Membrane Binding Properties of the Caspase Generated Fragments of the Beaded Filament Structural Protein 1 (BFSP1) Involves an Amphipathic Helix. Cells, 12(12), Article 1580. https://doi.org/10.3390/cells12121580
- AmiP from hyperthermophilic Thermus parvatiensis prophage is a thermoactive and ultrathermostable peptidoglycan lytic amidaseJasilionis, A., Plotka, M., Wang, L., Dorawa, S., Lange, J., Watzlawick, H., van den Bergh, T., Vroling, B., Altenbuchner, J., Kaczorowska, A., Pohl, E., Kaczorowski, T., Nordberg Karlsson, E., & Freitag‐Pohl, S. (2023). AmiP from hyperthermophilic Thermus parvatiensis prophage is a thermoactive and ultrathermostable peptidoglycan lytic amidase. Protein Science, 32(3), Article e4585. https://doi.org/10.1002/pro.4585
- Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasisSowerby, K., Freitag-Pohl, S., Murillo, A. M., Silber, A. M., & Pohl, E. (2023). Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasis. Acta Crystallographica Section D Structural Biology, 79(6), 518-530. https://doi.org/10.1107/s2059798323003613
- CPR-C4 is a highly conserved novel protease from the Candidate Phyla Radiation with remote structural homology to human vasohibinsCornish, K. A., Lange, J., Aevarsson, A., & Pohl, E. (2022). CPR-C4 is a highly conserved novel protease from the Candidate Phyla Radiation with remote structural homology to human vasohibins. Journal of Biological Chemistry, 298(5), Article 101919. https://doi.org/10.1016/j.jbc.2022.101919
- Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistanceSchwarz, M., Eno, R. F., Freitag-Pohl, S., Coxon, C. R., Straker, H. E., Wortley, D. J., Hughes, D. J., Mitchell, G., Moore, J., Cummins, I., Onkokesung, N., Brazier-Hicks, M., Edwards, R., Pohl, E., & Steel, P. G. (2021). Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance. Organic & Biomolecular Chemistry, 19(42), 9211-9222. https://doi.org/10.1039/d1ob01802g
- Multiple Occurrences of a 168-Nucleotide Deletion in SARS-CoV-2 ORF8, Unnoticed by Standard Amplicon Sequencing and Variant Calling PipelinesBrandt, D., Simunovic, M., Busche, T., Haak, M., Belmann, P., Jünemann, S., Schulz, T., Klages, L. J., Vinke, S., Beckstette, M., Pohl, E., Scherer, C., Sczyrba, A., & Kalinowski, J. (2021). Multiple Occurrences of a 168-Nucleotide Deletion in SARS-CoV-2 ORF8, Unnoticed by Standard Amplicon Sequencing and Variant Calling Pipelines. Viruses, 13(9). https://doi.org/10.3390/v13091870
- Structure-functional relationship of cellular retinoic acid binding proteins I and II interacting with natural and synthetic ligandsTomlinson, C. W., Cornish, K. A., Whiting, A., & Pohl, E. (2021). Structure-functional relationship of cellular retinoic acid binding proteins I and II interacting with natural and synthetic ligands. Acta Crystallographica. Section D, Structural Biology., 77(2), 164-175. https://doi.org/10.1107/s2059798320015247
- Chalcones identify cTXNPx as a potential antileishmanial drug targetEscrivani, D. O., Charlton, R. L., Caruso, M. B., Burle-Caldas, G. A., Borsodi, M. P. G., Zingali, R. B., Arruda-Costa, N., Palmeira-Mello, M. V., de Jesus, J. B., Souza, A. M., Abrahim-Vieira, B., Freitag-Pohl, S., Pohl, E., Denny, P. W., Rossi-Bergmann, B., & Steel, P. G. (2021). Chalcones identify cTXNPx as a potential antileishmanial drug target. PLOS Neglected Tropical Diseases, 15(11), Article e0009951. https://doi.org/10.1371/journal.pntd.0009951
- Crystal structure of the GDP-bound GTPase domain of Rab5a from Leishmania donovaniZohib, M., Maheshwari, D., Pal, R. K., Freitag-Pohl, S., Biswal, B. K., Pohl, E., & Arora, A. (2020). Crystal structure of the GDP-bound GTPase domain of Rab5a from Leishmania donovani. Acta Crystallographica Section F: Structural Biology Communications, 76(11), 544-556. https://doi.org/10.1107/s2053230x20013722
- Obtaining Tertiary Protein Structures by the ab Initio Interpretation of Small Angle X-ray Scattering DataPrior, C., Davies, O. R., Bruce, D., & Pohl, E. (2020). Obtaining Tertiary Protein Structures by the ab Initio Interpretation of Small Angle X-ray Scattering Data. Journal of Chemical Theory and Computation, 16(3), 1985-2001. https://doi.org/10.1021/acs.jctc.9b01010
- GSP4PDB: a web tool to visualize, search and explore protein-ligand structural patternsAngles, R., Arenas-Salinas, M., García, R., Reyes-Suarez, J. A., & Pohl, E. (2020). GSP4PDB: a web tool to visualize, search and explore protein-ligand structural patterns. BMC Bioinformatics, 21(S2), Article 85. https://doi.org/10.1186/s12859-020-3352-x
- Crystal structures of the Bacillus subtilis prophage lytic cassette proteins XepA and YomSFreitag-Pohl, S., Jasilionis, A., Håkansson, M., Svensson, L. A., Kovačič, R., Welin, M., Watzlawick, H., Wang, L., Altenbuchner, J., Płotka, M., Kaczorowska, A. K., Kaczorowski, T., Nordberg Karlsson, E., Al-Karadaghi, S., Walse, B., Aevarsson, A., & Pohl, E. (2019). Crystal structures of the Bacillus subtilis prophage lytic cassette proteins XepA and YomS. Acta Crystallographica. Section D, Structural Biology., 75(11), 1028-1039. https://doi.org/10.1107/s2059798319013330
- Relative Binding Energies Predict Crystallographic Binding Modes of Ethionamide Booster Lead CompoundsTatum, N., Duarte, F., Kamerlin, S., & Pohl, E. (2019). Relative Binding Energies Predict Crystallographic Binding Modes of Ethionamide Booster Lead Compounds. Journal of Physical Chemistry Letters, 10(9), 2244-2249. https://doi.org/10.1021/acs.jpclett.9b00741
- A unique dynamin-related protein is essential for mitochondrial fission in Toxoplasma gondiiMelatti, C., Pieperhoff, M., Lemgruber, L., Pohl, E., Sheiner, L., & Meissner, M. (2019). A unique dynamin-related protein is essential for mitochondrial fission in Toxoplasma gondii. PLoS Pathogens, 15(4), Article e1007512. https://doi.org/10.1371/journal.ppat.1007512
- Fluorescent retinoic acid analogues as probes for biochemical and intracellular characterization of retinoid signalling pathwaysChisholm, D., Tomlinson, C., Zhou, G., Holden, C., Affleck, V., Lamb, R., Newling, K., Ashton, P., Valentine, R., Redfern, C., Erostyak, J., Makkai, G., Ambler, C., Whiting, A., & Pohl, E. (2019). Fluorescent retinoic acid analogues as probes for biochemical and intracellular characterization of retinoid signalling pathways. ACS Chemical Biology, 14(3), 369-377. https://doi.org/10.1021/acschembio.8b00916
- How to Stabilize Protein: Stability Screens for Thermal Shift Assays and Nano Differential Scanning Fluorimetry in the Virus-X ProjectBruce, D., Cardew, E., Freitag-Pohl, S., & Pohl, E. (2019). How to Stabilize Protein: Stability Screens for Thermal Shift Assays and Nano Differential Scanning Fluorimetry in the Virus-X Project. Journal of Visualized Experiments, 144, Article e58666. https://doi.org/10.3791/58666
- A novel fluorescence competition assay for retinoic acid binding proteinsTomlinson, C. W., Chisholm, D. R., Valentine, R., Whiting, A., & Pohl, E. (2018). A novel fluorescence competition assay for retinoic acid binding proteins. ACS Medicinal Chemistry Letters, 9(12), 1297–1300-1300. https://doi.org/10.1021/acsmedchemlett.8b00420
- The Calcium-dependent protein kinase 1 from Toxoplasma gondii as target for structure-based drug designCardew, E., Verlinde, C., & Pohl, E. (2018). The Calcium-dependent protein kinase 1 from Toxoplasma gondii as target for structure-based drug design. Parasitology, 145(2), 210-218. https://doi.org/10.1017/s0031182017001901
- Air/Liquid Interfacial Nanoassembly of Molecular Building Blocks into Preferentially Oriented Porous Organic Nanosheet Crystals via Hydrogen BondingMakiura, R., Tsuchiyama, K., Pohl, E., Prassides, K., Sakata, O., Tajiri, H., & Konovalov, O. (2017). Air/Liquid Interfacial Nanoassembly of Molecular Building Blocks into Preferentially Oriented Porous Organic Nanosheet Crystals via Hydrogen Bonding. ACS Nano, 11(11), 10875-10882. https://doi.org/10.1021/acsnano.7b04447
- New active leads for Tuberculosis booster drugs by structure-based drug discoveryTatum, N. J., Liebeschuezt, J., Cole, J. C., Frita, R., Herledan, A., Baulard, A., Willand, N., & Pohl, E. (2017). New active leads for Tuberculosis booster drugs by structure-based drug discovery. Organic and Biomolecular Chemistry, 15(48), 10245-10255. https://doi.org/10.1039/c7ob00910k
- Functional and phylogenetic evidence of a bacterial origin for the first enzyme in sphingolipid biosynthesis in a phylum of eukaryotic protozoan parasitesMina, J., Thye, J., Alqaisi, A., Bird, L., Dods, R., Groftehauge, M., Mosely, J., Pratt, S., Shams-Eldin, H., Schwarz, R., Pohl, E., & Denny, P. W. (2017). Functional and phylogenetic evidence of a bacterial origin for the first enzyme in sphingolipid biosynthesis in a phylum of eukaryotic protozoan parasites. Journal of Biological Chemistry, 292(29), 12208-12219. https://doi.org/10.1074/jbc.m117.792374
- A tight tunable range for Ni(II) sensing and buffering in cellsFoster, A., Pernil, R., Patterson, C., Scott, A., Pålsson, L., Pal, R., Cummins, I., Chivers, P., Pohl, E., & Robinson, N. (2017). A tight tunable range for Ni(II) sensing and buffering in cells. Nature Chemical Biology, 13(4), 409-414. https://doi.org/10.1038/nchembio.2310
- The molecular basis of the interactions between synthetic retinoic acid analogues and the retinoic acid receptorsHaffez, H., Chisholm, D., Valentine, R., Pohl, E., Redfern, C., & Whiting, A. (2017). The molecular basis of the interactions between synthetic retinoic acid analogues and the retinoic acid receptors. MedChemComm., 8(3), 578-592. https://doi.org/10.1039/c6md00680a
- Mutantelec: AnIn Silicomutation simulation platform for comparative electrostatic potential profiling of proteinsValdebenito-Maturana, B., Reyes-Suarez, J. A., Henriquez, J., Holmes, D. S., Quatrini, R., Pohl, E., & Arenas-Salinas, M. (2017). Mutantelec: AnIn Silicomutation simulation platform for comparative electrostatic potential profiling of proteins. Journal of Computational Chemistry, 38(7), 467-474. https://doi.org/10.1002/jcc.24712
- Conjugate Addition of 3-Buytn-2-one to Anilines in Ethanol: Alkene Geometric Insights through In Situ FTIR MonitoringChisholm, D. R., Valentine, R., Pohl, E., & Whiting, A. (2016). Conjugate Addition of 3-Buytn-2-one to Anilines in Ethanol: Alkene Geometric Insights through In Situ FTIR Monitoring. Journal of Organic Chemistry, 81(17), 7557-7565. https://doi.org/10.1021/acs.joc.6b01110
- The Effectors and Sensory Sites of Formaldehyde-Responsive Regulator FrmR and Metal-Sensing VariantOsman, D., Piergentili, C., Chen, J., Sayer, L., Usón, I., Huggins, T., Robinson, N., & Pohl, E. (2016). The Effectors and Sensory Sites of Formaldehyde-Responsive Regulator FrmR and Metal-Sensing Variant. Journal of Biological Chemistry, 291(37), 19502-19516. https://doi.org/10.1074/jbc.m116.745174
- Practical synthetic strategies towards lipophilic 6-iodotetrahydroquinolines and -dihydroquinolinesChisholm, D., Zhou, G., Pohl, E., Valentine, R., & Whiting, A. (2016). Practical synthetic strategies towards lipophilic 6-iodotetrahydroquinolines and -dihydroquinolines. Beilstein Journal of Organic Chemistry, 12, 1851-1862. https://doi.org/10.3762/bjoc.12.174
- The role of protein-ligand contacts in allosteric regulation of the Escherichia coli Catabolite Activator ProteinTownsend, P., Rodgers, T., Glover, L., Korhonen, H., Richards, S., Colwell, L., Pohl, E., Wilson, M., Hodgson, D., McLeish, T., & Cann, M. (2015). The role of protein-ligand contacts in allosteric regulation of the Escherichia coli Catabolite Activator Protein. Journal of Biological Chemistry, 290(36), 22225-22235. https://doi.org/10.1074/jbc.m115.669267
- Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide AdductGrøftehauge, M., Truan, D., Vasil, A., Denny, P., Vasil, M., & Pohl, E. (2015). Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct. International Journal of Molecular Sciences, 16(7), 15971-15984. https://doi.org/10.3390/ijms160715971
- Global low-frequency motions in protein allostery: CAP as a model systemTownsend, P., Rogers, T., Pohl, E., Wilson, M., McLeish, T., & Cann, M. (2015). Global low-frequency motions in protein allostery: CAP as a model system. Biophysical Reviews, 7(2), 175-182. https://doi.org/10.1007/s12551-015-0163-9
- A silk purse from a sow’s ear – bioinspired materials based on α-helical coiled coilsQuinlan, R., Bromley, E., & Pohl, E. (2015). A silk purse from a sow’s ear – bioinspired materials based on α-helical coiled coils. Current Opinion in Cell Biology, 32, 131-137. https://doi.org/10.1016/j.ceb.2014.12.010
- Protein–ligand interactions investigated by thermal shift assays (TSA) and dual polarization interferometry (DPI)Grøftehauge, M. K., Hajizadeh, N. R., Swann, M. J., & Pohl, E. (2015). Protein–ligand interactions investigated by thermal shift assays (TSA) and dual polarization interferometry (DPI). Acta Crystallographica . Section D, Biological Crystallography, 71(1), 36-44. https://doi.org/10.1107/s1399004714016617
- The crystal structures of apo and cAMP-bound GlxR from Corynebacterium glutamicum reveal structural and dynamic changes upon cAMP binding in CRP/FNR family transcription factorsTownsend, P., Jungwirth, B., Pojer, P., Bußmann, M., Money, V., Cole, S., Pühler, A., Tauch, A., Bott, M., Cann, M., & Pohl, E. (2014). The crystal structures of apo and cAMP-bound GlxR from Corynebacterium glutamicum reveal structural and dynamic changes upon cAMP binding in CRP/FNR family transcription factors. PLoS ONE, 9(12), Article e113265. https://doi.org/10.1371/journal.pone.0113265
- The crystal structure of ferritin from Chlorobium tepidum reveals a new conformation of the 4-fold channel for this protein familyArenas-Salinas, M., Townsend, P., Brito, C., Marquez, V., Marabolli, V., Gonzalez-Nilo, F., Matias, C., Watt, R., López-Castro, J., Domínguez-Vera, J., Pohl, E., & Yévenes, A. (2014). The crystal structure of ferritin from Chlorobium tepidum reveals a new conformation of the 4-fold channel for this protein family. Biochimie, 106, 39-47. https://doi.org/10.1016/j.biochi.2014.07.019
- Structure solution of DNA-binding proteins and complexes with ARCIM- BOLDO librariesPröpper, K., Meindl, K., Sammito, M., Dittrich, B., Sheldrick, G., Pohl, E., & Uson, I. (2014). Structure solution of DNA-binding proteins and complexes with ARCIM- BOLDO libraries. Acta Crystallographica. Section D, Biological Crystallography., 70(6), 1743-1757. https://doi.org/10.1107/s1399004714007603
- High-level over-expression, purification, and crystallization of a novel phospholipase C/sphingomyelinase from Pseudomonas aeruginosaTruan, D., Vasil, A., Stonehouse, M., Vasil, M., & Pohl, E. (2013). High-level over-expression, purification, and crystallization of a novel phospholipase C/sphingomyelinase from Pseudomonas aeruginosa. Protein Expression and Purification, 90(1), 40-46. https://doi.org/10.1016/j.pep.2012.11.005
- Changes in the quaternary structure and function of MjHSP16.5 attributable to deletion of the I–X–I motif and introduction of the substitution, R107G in the a-crystallin domainQuinlan, R., Zhang, Y., Lansbury, A., Williamson, I., Pohl, E., & Sun, F. (2013). Changes in the quaternary structure and function of MjHSP16.5 attributable to deletion of the I–X–I motif and introduction of the substitution, R107G in the a-crystallin domain. Philosophical Transactions of the Royal Society B: Biological Sciences, 368(1617), Article 20120327. https://doi.org/10.1098/rstb.2012.0327
- A Nucleotide Phosphatase Activity in the Nucleotide Binding Domain of an Orphan Resistance Protein from RiceFenyk, S., de San Eustaquio Campillo, A., Pohl, E., Hussey, P., & Cann, M. (2012). A Nucleotide Phosphatase Activity in the Nucleotide Binding Domain of an Orphan Resistance Protein from Rice. Journal of Biological Chemistry, 287(6), 4023-4032. https://doi.org/10.1074/jbc.m111.314450
- Synthesis and molecular structure of a perfluorinated pyridyl carbanionColgin, N., Tatum, N., Pohl, E., Cobb, S., & Sandford, G. (2012). Synthesis and molecular structure of a perfluorinated pyridyl carbanion. Journal of Fluorine Chemistry, 133, 33-37. https://doi.org/10.1016/j.jfluchem.2011.09.013
- Metal-adeninate vertices for the construction of an exceptionally porous metal-organic frameworkAn, J., Farha, O. K., Hupp, J. T., Pohl, E., Yeh, J. I., & Rosi, N. L. (2012). Metal-adeninate vertices for the construction of an exceptionally porous metal-organic framework. Nature Communications, 3, Article 604. https://doi.org/10.1038/ncomms1618
- The Crystal Structure of Non-Modified and Bipyridine-Modified PNA DuplexesYeh, J. I., Pohl, E., Truan, D., He, W., Sheldrick, G. M., Du, S., & Achim, C. (2010). The Crystal Structure of Non-Modified and Bipyridine-Modified PNA Duplexes. Chemistry - A European Journal, 16(39), 11867-11875. https://doi.org/10.1002/chem.201000392
- Crystal Structure of the Caseinolytic Protease Gene Regulator, a Transcriptional Activator in ActinomycetesRusso, S., Schweitzer, J., Polen, T., Bott, M., & Pohl, E. (2009). Crystal Structure of the Caseinolytic Protease Gene Regulator, a Transcriptional Activator in Actinomycetes. Journal of Biological Chemistry, 284(8), 5208-5216. https://doi.org/10.1074/jbc.m806591200
- The Metal-Dependent Regulators FurA and FurB from Mycobacterium TuberculosisLucarelli, D., Vasil, M., Meyer-Klaucke, W., & Pohl, E. (2008). The Metal-Dependent Regulators FurA and FurB from Mycobacterium Tuberculosis. International Journal of Molecular Sciences, 9(8), 1548-1560. https://doi.org/10.3390/ijms9081548
- Crystallization and preliminary X-ray analysis of the Thermoplasma acidophilum 20S proteasome in complex with protein substrates.Felderer, K., Groves, M., Diez, J., Pohl, E., & Witt, S. (2008). Crystallization and preliminary X-ray analysis of the Thermoplasma acidophilum 20S proteasome in complex with protein substrates. Acta Crystallographica. Section F, Structural Biology Communications., 64(10), 899-902. https://doi.org/10.1107/s1744309108026791
- Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosisLucarelli, D., Russo, S., Garman, E., Milano, A., Meyer-Klaucke, W., & Pohl, E. (2007). Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. Journal of Biological Chemistry, 282(13), 9914-9922. https://doi.org/10.1074/jbc.m609974200
- Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulatorPohl, E., Haller, J., Mijovilovich, A., Meyer-Klaucke, W., Garman, E., & Vasil, M. (2003). Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Molecular Microbiology, 47(4), 903-915. https://doi.org/10.1046/j.1365-2958.2003.03337.x

