Staff profile

| Affiliation |
|---|
| Professor in the Department of Biosciences |
| Visiting Associate Professor in the Department of Chemistry |
| Professor in the Biophysical Sciences Institute |
| Biophysical Sciences Institute Executive Board in the Biophysical Sciences Institute |
Biography
Toxin-antitoxin systems and bacteriophage-resistance
Whilst bacteria are often thought of as selfish cells working for their own benefit, we can observe that they exist as diverse interacting communities. This is reflected in the ubiquitous presence and implementation of "toxin-antitoxin" systems throughout known Bacterial and Archaeal species. Toxin-antitoxin systems are characterised as small genetic loci generally encoding two parts. The toxin, when free to act, will target the host cell and stall growth. In the presence of the antitoxin, this effect is negated and cells grow freely.
It might appear peculiar that bacterial cells carry toxin-antitoxin systems, until you consider the potential advantages. For instance, if there aren't enough nutrients to go around, one cell activates its internal toxins, allowing it to grow slower or die, so that the population of clonal bacteria around it can survive. Another example would be when a bacterial cell becomes infected by a bacteria-specific virus, called a bacteriophage (phage). Unchecked, the phage would replicate, burst out, and infect neighbour cells. If the infected cell shuts down quickly, however, it can stop viral spread and protect the bacterial population.
Toxins from toxin-antitoxin systems often target the same biomolecules as antibiotics. Studying how these toxins kill bacteria will allow us to develop new ideas and methods for stopping bacterial infections.
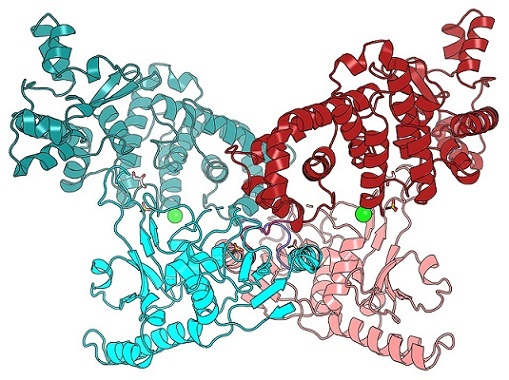
Figure 1. The BrxU DNA-modification dependent restriction enzyme targets modifed phage DNA for cleavage. This crystal structure shows an intertwined BrxU dimer, with each monomer coloured in cyans or pinks. See Picton et al. 2021, Nucleic Acids Research 49(19): 11257-11273.
Harnessing molecular tools from phage-host interactions
The interactions between phages and their bacterial hosts have generated a wealth of tools used in biotechnology, including CRISPR-Cas systems and restriction enzymes (Figure. 1).
As the natural predators of bacteria, it is also essential to investigate phage-host interactions in order to develop phages as a viable therapeutic alternative to antibiotics.
We investigate toxin-antitoxin systems and phage-host interactions using a range of molecular biology, microbiology, genetic and biochemical techniques. These include protein biochemistry, genomics and structural analysis through X-ray crystallography.
Research interests
- Antimicrobial Resistance
- Bacteriophage biology
- Biochemistry
- Molecular Microbiology
- Structural Biology
- Toxin-antitoxin systems
- Phage defence
Esteem Indicators
- 2019: Lister Institute Prize Fellow:
- 2017: Springboard Award:
Publications
Journal Article
- Blower, T. R., & van Houte, S. (2024). Viruses wrap up bacterial defence systems. Nature, 625, 250-251. https://doi.org/10.1038/d41586-023-03796-8
- Xu, X., Barriot, R., Voisin, B., Arrowsmith, T. J., Usher, B., Gutierrez, C., Han, X., Pagès, C., Redder, P., Blower, T. R., Neyrolles, O., & Genevaux, P. (2024). Nucleotidyltransferase toxin MenT extends aminoacyl acceptor ends of serine tRNAs to control Mycobacterium tuberculosis growth. Nature Communications, 15(1), Article 9596. https://doi.org/10.1038/s41467-024-53931-w
- Arrowsmith, T. J., Xu, X., Xu, S., Usher, B., Stokes, P., Guest, M., Bronowska, A. K., Genevaux, P., & Blower, T. R. (2024). Inducible auto-phosphorylation regulates a widespread family of nucleotidyltransferase toxins. Nature Communications, 15(1), Article 7719. https://doi.org/10.1038/s41467-024-51934-1
- Went, S. C., Picton, D. M., Morgan, R. D., Nelson, A., Brady, A., Mariano, G., Dryden, D. T. F., Smith, D. L., Wenner, N., Hinton, J. C. D., & Blower, T. R. (2024). Structure and rational engineering of the PglX methyltransferase and specificity factor for BREX phage defence. Nature Communications, 15, Article 7236. https://doi.org/10.1038/s41467-024-51629-7
- Birkholz, N., Kamata, K., Feussner, M., Wilkinson, M. E., Cuba Samaniego, C., Migur, A., Kimanius, D., Ceelen, M., Went, S. C., Usher, B., Blower, T. R., Brown, C. M., Beisel, C. L., Weinberg, Z., Fagerlund, R. D., Jackson, S. A., & Fineran, P. C. (2024). Phage anti-CRISPR control by an RNA- and DNA-binding helix–turn–helix protein. Nature, 631(8021), 670-677. https://doi.org/10.1038/s41586-024-07644-1
- Ridgway, R., Lu, H., Blower, T. R., Evans, N. J., & Ainsworth, S. (2024). Genomic and taxonomic evaluation of 38 Treponema prophage sequences. BMC Genomics, 25(1), Article 549. https://doi.org/10.1186/s12864-024-10461-5
- Agapov, A., Baker, K., Bedekar, P., Bhatia, R., Blower, T. R., Brockhurst, M. A., Brown, C., Chong, C., Fothergill, J. L., Graham, S., Hall, J. P. J., Maestri, A., McQuarrie, S., Olina, A., Pagliara, S., Recker, M., Richmond, A., Shaw, S. J., Szczelkun, M. D., Taylor, T. B., …Wright, R. (2024). Multi-layered genome defences in bacteria. Current Opinion in Microbiology, 78, Article 102436. https://doi.org/10.1016/j.mib.2024.102436
- Beck, I. N., Arrowsmith, T. J., Grobbelaar, M. J., Bromley, E. C., Marles-Wright, J., & Blower, T. R. (2024). Toxin release by conditional remodelling of ParDE1 from Mycobacterium tuberculosis leads to gyrase inhibition. Nucleic Acids Research, 52(4), 1909-1929. https://doi.org/10.1093/nar/gkad1220
- Bandak, A. F., Blower, T. R., Nitiss, K. C., Shah, V., Nitiss, J., & Berger, J. (2024). Using energy to go downhill—a genoprotective role for ATPase activity in DNA topoisomerase II. Nucleic Acids Research, 52(3), 1313–1324. https://doi.org/10.1093/nar/gkad1157
- Kelly, A., Went, S. C., Mariano, G., Shaw, L. P., Picton, D. M., Duffner, S. J., Coates, I., Herdman-Grant, R., Gordeeva, J., Drobiazko, A., Isaev, A., Lee, Y.-J., Luyten, Y., Morgan, R. D., Weigele, P., Severinov, K., Wenner, N., Hinton, J. C., & Blower, T. R. (2023). Diverse Durham collection phages demonstrate complex BREX defence responses. Applied and Environmental Microbiology, 89(9), Article e00623-23. https://doi.org/10.1128/aem.00623-23
- Bandak, A., Blower, T., Nitiss, K., Gupta, R., Lau, A., Guha, R., Nitiss, J., & Berger, J. (2023). Naturally mutagenic sequence diversity in a human type II topoisomerase. Proceedings of the National Academy of Sciences, 120(28), Article e2302064120. https://doi.org/10.1073/pnas.2302064120
- Mariano, G., & Blower, T. (2023). Conserved domains can be found across distinct phage defence systems. Molecular Microbiology, 120(1), 45-53. https://doi.org/10.1111/mmi.15047
- Macdonald, E., Wright, R., Connolly, J., Strahl, H., Brockhurst, M., van Houte, S., Blower, T., Palmer, T., & Mariano, G. (2023). The novel anti-phage system Shield co-opts an RmuC domain to mediate phage defense across Pseudomonas species. PLoS Genetics, 19(6), Article e1010784. https://doi.org/10.1371/journal.pgen.1010784
- Kelly, A., Arrowsmith, T., Went, S., & Blower, T. (2023). Toxin–antitoxin systems as mediators of phage defence and the implications for abortive infection. Current Opinion in Microbiology, 73, Article 102293. https://doi.org/10.1016/j.mib.2023.102293
- Xu, X., Usher, B., Gutierrez, C., Barriot, R., Arrowsmith, T. J., Han, X., Redder, P., Neyrolles, O., Blower, T. R., & Genevaux, P. (2023). MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding. Nature Communications, 14, Article 4644. https://doi.org/10.1038/s41467-023-40264-3
- Ling, E., Baslé, A., Cowell, I., Van Den Berg, B., Blower, T., & Austin, C. (2022). A comprehensive structural analysis of the ATPase domain of Human DNA topoisomerase II Beta bound to AMPPNP, ADP and the bisdioxopiperazine, ICRF193. Structure, 30(8), P1129-1145.e3. https://doi.org/10.1016/j.str.2022.05.009
- Beck, I., Picton, D., & Blower, T. (2022). Crystal structure of the BREX phage defence protein BrxA. Current Research in Structural Biology, 4, 211-219. https://doi.org/10.1016/j.crstbi.2022.06.001
- Picton, D., Harling-Lee, J., Duffner, S., Went, S., Morgan, R., Hinton, J., & Blower, T. (2022). A widespread family of WYL-domain transcriptional regulators co-localises with diverse phage defence systems and islands. Nucleic Acids Research, 50(9), 5191-5207. https://doi.org/10.1093/nar/gkac334
- Guillen-Garcia, A., Gibson, S., Jordan, C., Ramaswamy, V., Linthwaite, V., Bromley, E., Brown, A., Hodgson, D., Blower, T., Verlet, J., Degiacomi, M., Palsson, L.-O., & Cann, M. (2022). Allophycocyanin A is a carbon dioxide receptor in the cyanobacterial phycobilisome. Nature Communications, 13, Article 5289. https://doi.org/10.1038/s41467-022-32925-6
- Picton, D., Luyten, Y., Morgan, R., Nelson, A., Smith, D., Dryden, D., Hinton, J., & Blower, T. (2021). The phage defence island of a multidrug resistant plasmid uses both BREX and type IV restriction for complementary protection from viruses. Nucleic Acids Research, 49(19), 11257-11273. https://doi.org/10.1093/nar/gkab906
- Usher, B., Birkholz, N., Beck, I., Fagerlund, R., Jackson, S., Fineran, P., & Blower, T. (2021). Crystal structure of the anti-CRISPR repressor Aca2. Journal of Structural Biology, 213(3), Article 107752. https://doi.org/10.1016/j.jsb.2021.107752
- Rodwell, E., Wenner, N., Pulford, C., Cai, Y., Bowers-Barnard, A., Beckett, A., Rigby, J., Picton, D., Blower, T., Feasey, N., Hinton, J., & Perez-Sepulveda, B. (2021). Isolation and characterisation of bacteriophages with activity against invasive non-typhoidal Salmonella causing blood-stream infection in Malawi. Viruses, 13(3), Article 478. https://doi.org/10.3390/v13030478
- Cai, Y., Usher, B., Gutierrez, C., Tolcan, A., Mansour, M., Fineran, P., Condon, C., Neyrolles, O., Genevaux, P., & Blower, T. (2020). A nucleotidyltransferase toxin inhibits growth of Mycobacterium tuberculosis through inactivation of tRNA acceptor stems. Science Advances, 6(31), Article eabb6651. https://doi.org/10.1126/sciadv.abb6651
- Beck, I., Usher, B., Hampton, H., Fineran, P., & Blower, T. (2020). Antitoxin autoregulation of M. tuberculosis toxin-antitoxin expression through negative cooperativity arising from multiple inverted repeat sequences. Biochemical Journal, 477(12), 2401-2419. https://doi.org/10.1042/bcj20200368
- Cross, J., Blower, T., Kingdon, A., Pal, R., Picton, D., & Walton, J. (2020). Anticancer Ruthenium Complexes with HDAC Isoform Selectivity. Molecules, 25(10), Article 2383. https://doi.org/10.3390/molecules25102383
- Blower, T., Bandak, A., Lee, A., Austin, C., Nitiss, J., & Berger, J. (2019). A complex suite of loci and elements in eukaryotic type II topoisomerases determine selective sensitivity to distinct poisoning agents. Nucleic Acids Research, 47(15), 8163-8179. https://doi.org/10.1093/nar/gkz579
- Gibson, E., Blower, T., Cacho, M., Bax, B., Berger, J., & Osheroff, N. (2018). Mechanism of Action of Mycobacterium tuberculosis Gyrase Inhibitors: A Novel Class of Gyrase Poisons. ACS Infectious Diseases, 4(8), 1211-1222. https://doi.org/10.1021/acsinfecdis.8b00035
- Hampton, H., Jackson, S., Fagerlund, R., Vogel, A., Dy, R., Blower, T., & Fineran, P. (2018). AbiEi binds cooperatively to the Type IV abiE toxin-antitoxin operator via a positively-charged surface and causes DNA bending and negative autoregulation. Journal of Molecular Biology, 430(8), 1141-1156. https://doi.org/10.1016/j.jmb.2018.02.022
- Ashley, R., Blower, T., Berger, J., & Osheroff, N. (2017). Recognition of DNA Supercoil Geometry by Mycobacterium tuberculosis Gyrase. Biochemistry, 56(40), 5440-5448. https://doi.org/10.1021/acs.biochem.7b00681
- Blower, T., Chai, R., Przybilski, R., Chindhy, S., Fang, X., Kidman, S., Tan, H., Luisi, B., Fineran, P., & Salmond, G. (2017). Evolution of Pectobacterium bacteriophage ΦM1 to escape two bifunctional Type III toxin-antitoxin and abortive infection systems through mutations in a single viral gene. Applied and Environmental Microbiology, 83(8), e03229-16. https://doi.org/10.1128/aem.03229-16
- Cross, J., Blower, T., Gallagher, N., Gill, J., Rockley, K., & Walton, J. (2016). Anticancer RuII and RhIII Piano-Stool Complexes that are Histone Deacetylase Inhibitors. ChemPlusChem, 81(12), 1276-1280. https://doi.org/10.1002/cplu.201600413
- Aldred, K., Blower, T., Kerns, R., Berger, J., & Osheroff, N. (2016). Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: Enhancing drug activity against wild-type and resistant gyrase. Proceedings of the National Academy of Sciences, 113(7), E839-E846. https://doi.org/10.1073/pnas.1525055113
- Blower, T., Williamson, B., Kerns, R., & Berger, J. (2016). Crystal structure and stability of gyrase–fluoroquinolone cleaved complexes from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences, 113(7), 1706-1713. https://doi.org/10.1073/pnas.1525047113
- Rao, F., Short, F., Voss, J., Blower, T., Orme, A., Whittaker, T., Luisi, B., & Salmond, G. (2015). Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein–RNA toxin–antitoxin system. Nucleic Acids Research, 43(19), 9529-9540. https://doi.org/10.1093/nar/gkv868
- Unwin, R., Cabanas, R., Yanagishima, T., Blower, T., Takahashi, H., Salmond, G., Edwardson, J., Fraden, S., & Eiser, E. (2015). DNA driven self-assembly of micron-sized rods using DNA-grafted bacteriophage fd virions. Physical Chemistry Chemical Physics, 17(12), 8194-202. https://doi.org/10.1039/c4cp05405a
- Short, F., Pei, X., Blower, T., Ong, S., Fineran, P., Luisi, B., & Salmond, G. (2013). Selectivity and self-assembly in the control of a bacterial toxin by an antitoxic noncoding RNA pseudoknot. Proceedings of the National Academy of Sciences, 110(3), E241-9. https://doi.org/10.1073/pnas.1216039110
- Blower, T., Short, F., Fineran, P., & Salmond, G. (2012). Viral molecular mimicry circumvents abortive infection and suppresses bacterial suicide to make hosts permissive for replication. Bacteriophage, 2(4), 234-238. https://doi.org/10.4161/bact.23830
- Short, F., Blower, T., & Salmond, G. (2012). A promiscuous antitoxin of bacteriophage T4 ensures successful viral replication. Molecular Microbiology, 83(4), 665-668. https://doi.org/10.1111/j.1365-2958.2012.07974.x
- Blower, T., Evans, T., Przybilski, R., Fineran, P., & Salmond, G. (2012). Viral Evasion of a Bacterial Suicide System by RNA-Based Molecular Mimicry Enables Infectious Altruism. PLoS Genetics, 8(10), https://doi.org/10.1371/journal.pgen.1003023
- Blower, T., Short, F., Rao, F., Mizuguchi, K., Pei, X., Fineran, P., Luisi, B., & Salmond, G. (2012). Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Research, 40(13), 6158-6173. https://doi.org/10.1093/nar/gks231
- Blower, T., Pei, X., Short, F., Fineran, P., Humphreys, D., Luisi, B., & Salmond, G. (2011). A processed non-coding RNA regulates an altruistic bacterial antiviral system. Nature Structural and Molecular Biology, 18(2), 185-190. https://doi.org/10.1038/nsmb.1981
- Blower, T., Salmond, G., & Luisi, B. (2011). Balancing at survival's edge: the structure and adaptive benefits of prokaryotic toxin-antitoxin partners. Current Opinion in Structural Biology, 21(1), 109-18. https://doi.org/10.1016/j.sbi.2010.10.009
- Blower, T., Fineran, P., Johnson, M., Toth, I., Humphreys, D., & Salmond, G. (2009). Mutagenesis and functional characterisation of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. Journal of Bacteriology, 191(19), 6029-6039. https://doi.org/10.1128/jb.00720-09
- Fineran, P., Blower, T., Foulds, I., Humphreys, D., Lilley, K., & Salmond, G. (2009). The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proceedings of the National Academy of Sciences, 106(3), 894-899. https://doi.org/10.1073/pnas.0808832106

