How do plants control lipid biosynthesis? The role of ACC1 pre-mRNA splicing
Background
The enzyme acetyl-CoA carboxylase (ACCase) is essential for fatty acid and lipid synthesis in plants, and exists in Arabidopsis as one plastidic heteromeric form and two homomeric cytosolic isoforms, ACC1 and ACC2. Plants that are mutant for the ACC1 gene show that this protein is essential for the synthesis of very long chain fatty acids (VLCFAs) and storage oils (triacyl glycerols, TAGs) in Arabidopsis, and interestingly is also required for correct patterning of cell division in the developing Arabidopsis embryo (acc1 mutants have been identified independently in different labs and named gurke and pasticcino3). While the ACC1 gene is transcribed in both seed and leaves, the accumulation of storage triacyl glycerols (TAGs) only occurs at high levels in the seed, suggesting differential post-transcriptional regulation of the gene/enzyme.
One mechanism that determines differential functionality of mRNAs is alternative splicing, whereby introns, or other sequences outside the coding region, such as in the 5' untranslated region (5'UTR), are differentially removed from the pre-mRNA to produce structurally different mature mRNAs. This can affect the functionality of the resultant protein translation product. In investigating the control of possible post-transcriptional control of ACC1 activity, we have evidence to suggest that the ACC1 transcript is alternatively spliced, with different splice isoforms present at different abundances in different tissues, potentially explaining the different ACC1 enzyme activities in different tissues. The ACC1 mRNA is also mis-spliced in the splicing mutant meristem-defective (mdf), which is defective in the splicing factor MDF, a homologue of both the human hSART1 and the yeast Snu66 SR proteins and spliceosome components.
Analysis of ACC1 splice isoforms in both wildtype seedlings and in seedlings of the mdf loss-of-function mutant, which is defective in splicing control, shows that the ACC1 transcript has an alternative donor site in the 5' UTR, upstream of the translation start site. The use of this alternative 5' UTR splice site in WT leaf and in the mdf mutant results in incomplete splicing, with the retention of an 'outron' (a retained intron outside the coding sequence; Fig. 1) which we hypothesise could affect mRNA stability or translation efficiency, and so ACC1 enzyme activity.
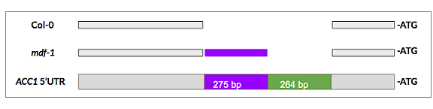
Fig. 1: 5'UTR regions of wildtype (Col-0) and mdf1 mutant splice variants, and (below) ACC1 unspliced 5'UTR.
Leaf and seed show different proportions of spliced and non-spliced versions of the ACC1 transcript, with the non-spliced isoform being relatively abundant in leaf and also in the mdf mutant, consistent with a role for splicing in regulating ACC1 activity (Fig. 2). Mis-splicing of ACC1 pre-mRNA in the mdf mutant or in transgenic MDF overexpressers is associated with ectopic lipid accumulation, defective membrane-dependent PIN protein localization and aberrant cell division in the root meristem. Genetic complementation of the mdf mutant with the wildtype ACC1 gene under the control of the MDF gene promoter leads to partial rescue of cell division patterning in the mdf mutant root.
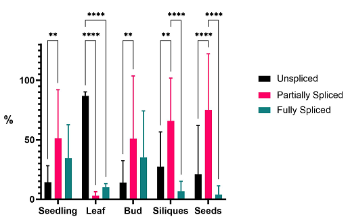
Fig. 2. Relative abundances of ACC1 5'UTR splice variants in Arabidopsis tissues.
These data show that correct splicing of ACC1 pre-mRNA is required for both correct cell division and tissue-specific oil accumulation in Arabidopsis.
Experimental approach
1. Further analysis of the splicing profile of ACC1 mRNA, using qRT-PCR in a) different wildtype seedling tissues (leaf, root, seed, flower) and b) mdf mutant tissues, to reveal tissue specificity of ACC1 splicing.
2. Measure ACC1 enzyme activity in different tissues of wiltype and mdf mutant, to determine a correlation between splice variant isoforms and enzyme functionality.
3. Further characterization of the proMDF::ACC1 transgenics in the mdf mutant background, to chacaterize the complementation phenotype. This will involve confocal microscopy imaging of the root tip, and root architecture measurements.
4. Further characterization of the lipid profiles, and PIN localization, of the proMDF::ACC1 complemented mdf mutant. This will involve lipid extraction and analysis by GC-MS, using the Department of Biosciences metabolomics facility.
The project will provide the student with experience in plant genetics, molecular biology, advanced bioimaging and biochemistry.
For more information, please contact Professor Keith Lindsey.


/prod01/prodbucket01/media/durham-university/departments-/biosciences/83453-1-1595X1594.jpg)